Projects

Environmental effects and dysregulation of T-cells in inflammatory diabetes
Type-1 Diabetes is an inflammatory autoimmune disease that develops during childhood where immune dysregulation leads to Beta-cell destruction and a loss of the bodies ability to regulate insulin production. This disease affects millions worldwide and is curiosly more common in highly modernized western countries. One of the most well known examples of this is the 6x greater risk of disease in finnish juveniles compared to russian juveniles. Currently, this is difficult to study due to high heterogenity in human populations. Our lab aims to develop both computational and in-vivo mouse models to better understand how environmental factors affect disease development.
We have recently published one of the first single cell RNA-seq studies in the translational NOD mouse model and found 1) High proportions of Double Negative T cells 2) Clonal specific regulation of phenotype in CD8+ T cells and 3) Immune metabolic dysregulation of pancreatic infiltrating T cells. To investigate these results further, our lab combines novel sequencing methods and in-vitro experimentation.

Determining the effects of diet on development of Colorectal cancer in higher-risk populations
Racial and genetic bias towards the development of cancer is a major factor that disproportionally impacts at-risk populations in the US. Black American populations are at higher risk of developing and dying from prostate and colorectal cancer. The higher incidence and mortality is not shared by native african populations who see equivalent risk to that of White American populations. Previous work by collaborator Rex Gaskins, found that this may be highly influenced by dietary factors including fats and carbohydrate intake, and physiological characteristics such as differences in bile acid production. We have recently conducted research showing that Black Americans produce more secondary bile acids and have a higher abundance of microbes associated with bile acid metabolism like C. scindens. We also found a negative relationship between omega-3 fatty acid intake and size of serrated polyps in Black Americans but not White americans. This has spurred further research by our lab to investigate the race specific effects of omega-3 and omega-6 fatty acids in serrated polypsosis.
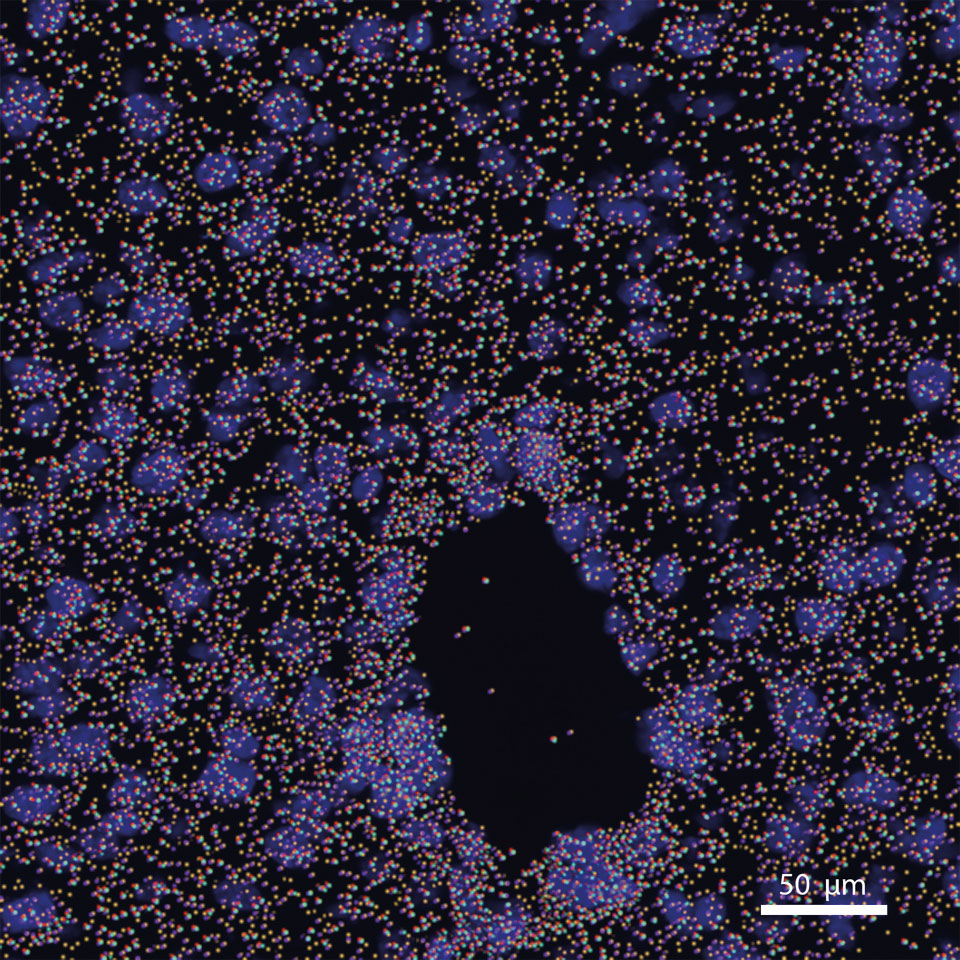
Computational and molecular method development for the understanding of immune interactions
Immune interactions between immune cells and non-immune represent the most complex tissue environment interactions in animals. Traditional in-vitro and in-vivo methods lack the resolution to fully capture the total possible interactions taking place in a disease context. Recent advances in sequencing, particularly in single cell and spatial technologies has enabled high throughput evaluation of the immune system that can capture the intricate interactions that underlie pathogenesis. In our lab we explore methodologies both computational and molecular that can improve the information that researchers can derive from these large scale sequencing experiments. Efforts in the lab include:
- Integrating and developing new techniques of single cell and spatial sequencing to reduce the cost for immune cell sequencing and analysis
- Developing new algorithms for immune phenotyping large scale sequencing data
- Developing machine learning methods for the classification of protein function in unannotated data
- Developing machine learning methods to predict TCR-pMHC interactions from large scale sequencing datasets
Packages
Coming soon…
Funding Sources
